Written by Grace Emin, DeNovix Application Scientist
Introduction
Nucleic acid quantitation relies on measuring the ultraviolet absorbance of DNA and RNA, most commonly at 260 nm, where the nucleobases absorb strongly. By applying the Beer-Lambert law together with extinction coefficients, absorbance values can be directly related to nucleic acid concentration. Absorbance at 230 nm and 260 nm provides a convenient basis for assessing sample purity, supporting quality control of genomic DNA, PCR products, and other prepared nucleic acid samples. Used alongside fluorescent dye-based assays, absorbance measurements form a practical framework for defining DNA or RNA concentration prior to downstream applications.
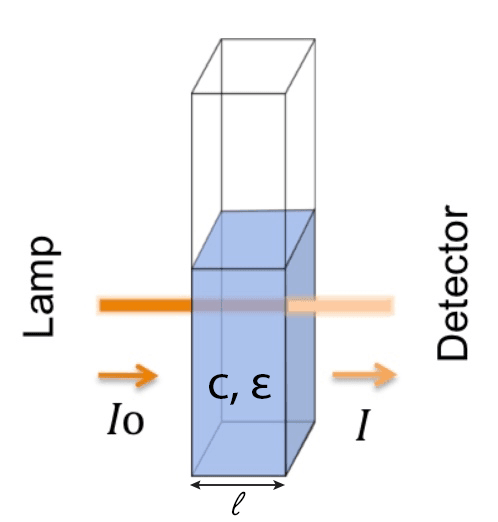
An understanding of Beer’s Law is needed to explore how concentration factors are calculated and used. Beer’s Law is an equation that describes the relationship between the concentration of a substance and its absorption of light at a known light path distance.
Beer’s Law Equation
Where:
- Abs = Absorbance
- l = Pathlength: The distance the light travels through the substance
- ε = Molar extinction coefficient: A measure of how strongly a substance absorbs light at a specific wavelength
- C = Concentration of the substance
The equation can be rearranged to solve for concentration:
In Beer’s Law, we can substitute the extinction coefficient with a nucleic acid concentration factor since it is another value that can be used to relate a substance’s ability to absorb light to its concentration. This can then be used for nucleic acid quantification.
What Are Nucleic Acid Concentration Factors, and How Are They Determined?
Nucleic acid concentration factors are wavelength-specific conversion factors that relate absorbance to concentration for different nucleic acid types, enabling direct calculation of DNA or RNA concentration from UV absorbance measurements.
These nucleic acid concentration factors were determined by measuring the concentration of a specific nucleic acid that is needed at a given pathlength to have an absorbance of one. It has been established that at a 1 cm path length, a concentration of 0.02 ng/μL of dsDNA was needed for an absorbance of one (1 Abs).
This concentration can then be used to solve for the factor (F) value by using the below rearranged Beer’s Law equation. The factor will then replace the extinction coefficient:
This factor can be used in the below rearranged Beer’s Law Equation to solve for an unknown concentration where the absorbance is measured, as done with the DS-11 Series Spectrophotometer:
The same calculations are used to determine the concentration factors of other nucleic acids. Other factors used by the DS-11 are below:
ssDNA: 33 ng/uL
ssRNA: 40 ng/uL
These concentration factors are inversely proportional to their extinction coefficient. The higher the quantity of nucleic acid required for 1A at 260 nm, the lower the corresponding concentration factor.
Why Are Nucleic Acid Concentration Factors Different?
dsDNA, ssDNA, and RNA have differing chemical and structural properties that result in varying light absorption behaviors. This translates to different concentrations of each nucleic acid required to result in 1.0 absorbance at 260 nm. The following characteristics of the nucleic acids contribute to the differing extinction coefficients and concentration factors:
- dsDNA: Its double helix structure, with the bonds being in close proximity to each other and the hydrogen bonds holding the structure together, results in a reduction in UV absorbance.
- ssDNA: Its bases are more exposed and not susceptible to the base stacking interactions like those observed with dsDNA.
- RNA: The ribose sugar and additional hydroxyl groups interfere with UV absorbance.
Learn How to Calculate the Concentration of Nucleic Acids with DeNovix
Nucleic acid concentration factors provide a simplified method for calculating nucleic acid concentrations using absorbance. The varying values appropriately consider the differing light absorption patterns as a result of the structural differences of the nucleic acids. You can check out the exceptional precision and linearity over a broad nucleic acid concentration range achieved by the DS-11 Series Spectrophotometer / Fluorometer by exploring our Microvolume Nucleic Acid Performance Data TechNote 105 found here.

Grace Emin
DeNovix Application Scientist
Grace Emin has been an Application Scientist at DeNovix Inc. since 2023. She graduated with a Master’s Degree in Biological Sciences with a Focus on Biomaterial Engineering and Disease Models. At DeNovix, Grace contributes to application and product development, as well as technical support, with expertise in imaging, assay development, and machine learning applications. When she’s not working, Grace enjoys painting, traveling, playing games, and spending time with her cat, Ivy.
Speak to a DeNovix Scientist
Have an application question, or want to learn more about our products? Click the button below to schedule a call with our team of expert application scientists!