The Nuclei AO/PI (left) and Trypan Blue Nuclei (right) apps on the CellDrop Automated Cell Counter are optimized with protocols for isolated nuclei counts.
Keep reading to learn how to count isolated nuclei on the CellDrop.


Nuclei Isolation Procedure & Considerations
Nuclei were isolated from HEK293T cell cultures according to the 10X Genomics® protocol for the “Isolation of Nuclei from Single Cell Suspensions. CG000124 Rev D.” Before lysis, cell density and viability were assessed using the standard CellDrop AO/PI protocol to confirm a minimum of ~2.5 million cells/mL at >90% viability.
With the basic performance established, isolations were performed from fresh and frozen tissue using Miltenyi Biotec’s Nuclei Extraction Buffer (130-128-024) and gentleMACs™ Dissociator. Images in this technical note have been selected from mouse liver samples, but extractions have been tested on mouse frozen mouse lung, prostate, kidney, brain, and liver, fresh mouse prostate, brain, and liver, and frozen rat lung.
Keeping the nuclei at 4°C throughout the protocol is important for high quality extractions. To ensure high yields, it is recommended to keep samples on fresh ice and work as quickly as possible. While not necessary for high yields, the gentleMACS™ Octo Coolers (130-130-533) were used in this procedure to keep nuclei at optimal temperatures throughout the extraction process.
The CellDrop count results showed a high extraction efficiency (>90%), but also contained cellular debris as is expected of whole organ isolations.
While the gentleMACs™ Dissociator system and filtration steps reduce debris, different tissue types may be more challenging to work with. Tissues from the brain or heart, for example, typically contain more debris than liver and lung samples.
An optional step is to remove debris from samples using kits such as the Miltenyi Anti-Nuclei Microbeads.
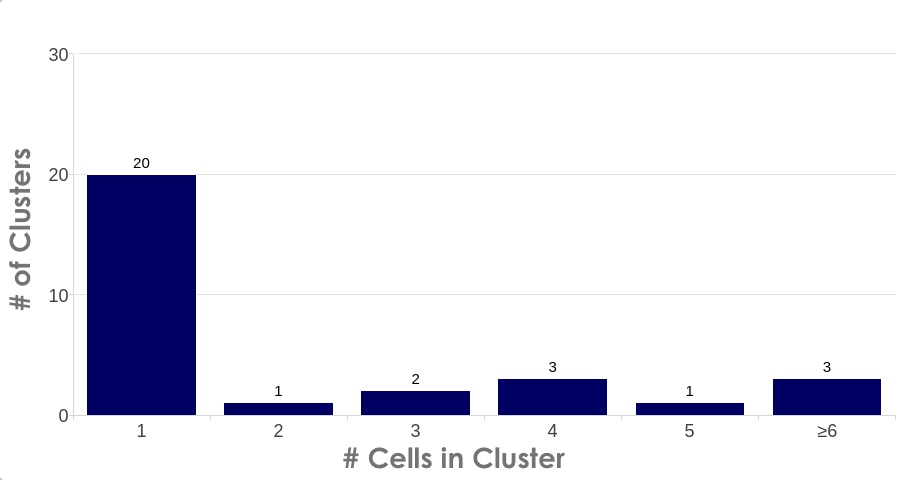
Minimizing debris and large clusters is important for the downstream workflow of single-cell sequencing. These can clog the fluidic chips, resulting in low quality libraries or failed sequencing experiments. Refer to the manufacturer’s protocol if large clusters of nuclei are observed. Similarly, removing intact cells that did not lyse during the procedure is also recommended.
CellDrop DirectPipette™ Technology
The unique DirectPipette™ technology of CellDrop Automated Cell Counters enables counting without disposable slides, which reduces plastic waste and costs. The variable chamber volume allows counting volumes of between 5 µL and 40 µL of sample.
In addition, CellDrop is also compatible with common disposable plastic or reusable slides to allow the user to both quantify the nuclear isolation on the CellDrop and transfer the same slide to a microscope with a higher magnification for nuclear integrity analysis.
Counting Nuclei with AO/PI Stain
Acridine Orange and Propidium Iodide (AO/PI) are used to determine the success of nuclei isolation. In traditional cell viability testing, the AO/PI dye combination stains live cells so they fluoresce green and dead cells fluoresce red. However, the stain will also label successfully isolated nuclei red and any remaining intact cells green. This allows the user to calculate the residual intact cells that carry over as a percent of the total counted and determine if the experimental workflow can proceed.
As the nuclear pore complex will allow passive diffusion up to 30-60 kDa, both AO and PI (~0.6 kDa) freely pass into the nucleus and display a red signal due to a FRET interaction between the two fluorophores. Minimizing the number of intact cells in isolation is important, so accurately enumerating the intact cells with AO/PI can improve quality control and improve consistency in the results of downstream workflows.
Software Results & Image Analysis
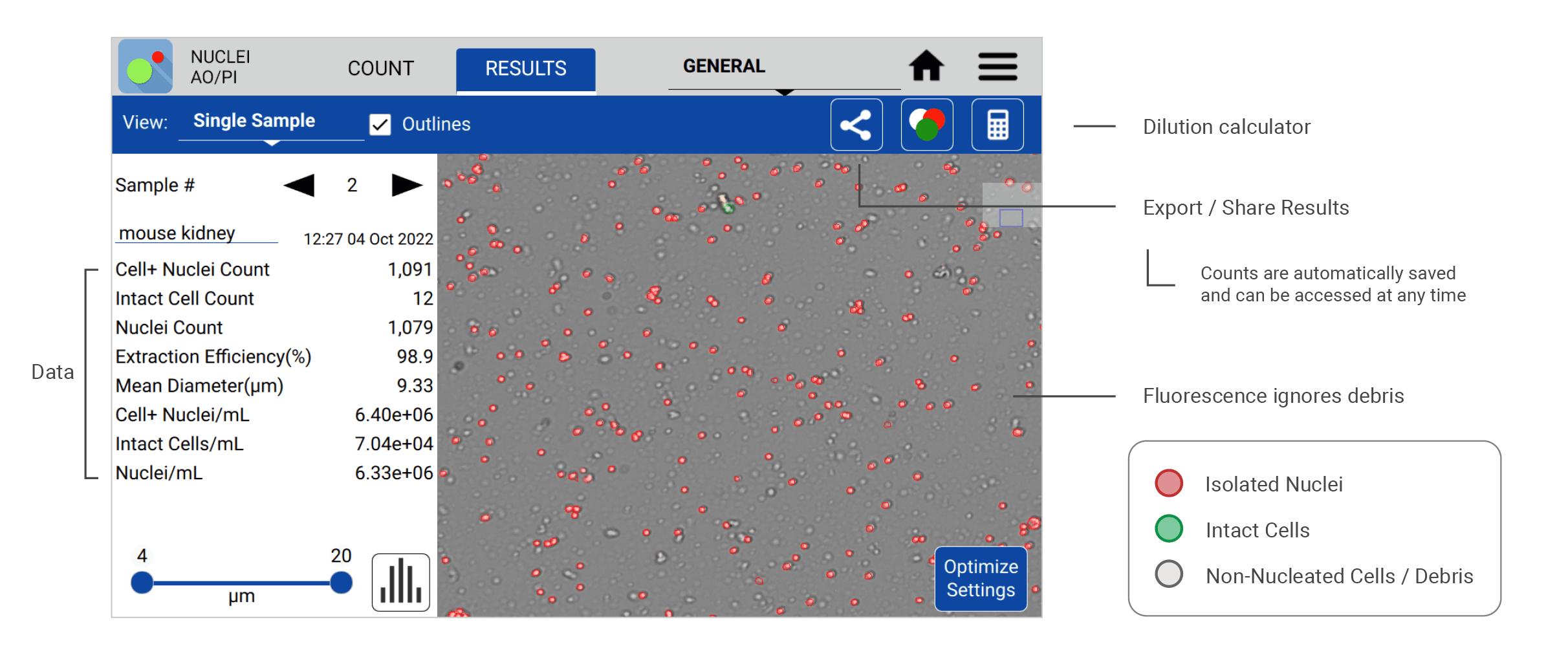
The following count (Figure 1) was made using the CellDrop Nuclei AO/PI App to analyze nuclei isolated from a mouse kidney sample. The CellDrop quantifies isolated nuclei (stained red) and the intact cells (stained green) to calculate the nuclei/mL concentration and nuclei extraction efficiency for each sample. Measurements are readily presented to the user on the results screen with other relevant sample data. Images and sample data are automatically saved to the instrument and can be recalled for review at any time.
A common requirement for high quality single nuclei sequencing library preparation is the need for cluster-free, monodispersed samples with accurate diameter assessment. Information such as nuclei/mL density, level of intact cell contamination via the extraction efficiency, and mean diameter size is readily accessible in the Results screen. Users can easily access advanced cluster analysis (Figure 2) on a per sample basis by switching to the Graphs view in the Results screen and selecting the Cluster Size bar graph by the dropdown menu.
Debris and Purification
The CellDrop fluorescence mode offers advantages, such as easily distinguishing background debris from nuclei, to increase the accuracy of results. However, large amounts of debris left in the sample might create challenges in downstream applications. Some tissue types may benefit from filtration or purification before moving onto the sequencing step.
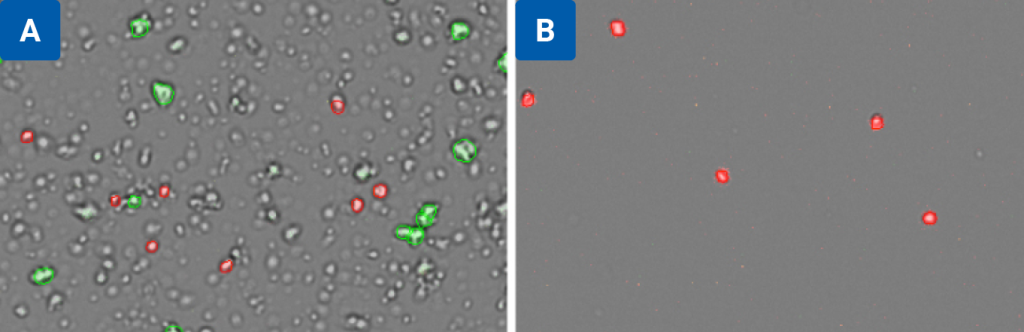
Figure 3 compares the same sample (isolated nuclei from mouse brain tissue) counted on the CellDrop Nuclei AO/PI App before (3A) and after (3B) purification. In 3A, the CellDrop software ignores debris and red blood cells to provide an accurate count of isolated nuclei and intact cells. In 3B, the same sample was analyzed again after nuclei enrichment using Miltenyi’s Anti-Nuclei Microbeads to remove debris.
Counting Nuclei with Trypan Blue Stain
Where dual fluorescence instrumentation is not available, it is also possible to analyze the success of a nuclear isolation using Trypan Blue. DeNovix recommends using fluorescent assays for quantifying isolated nuclei where possible due to the increased accuracy ensured by the clear differences in green and red signals. It should be noted that counting debris-laden samples using Trypan Blue can increase the number of erroneous counts either with an automated cell counter or by manual count.
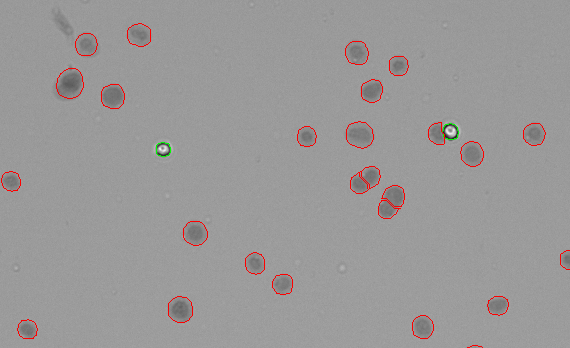
Figure 4 shows HEK293T cells stained with Trypan Blue and counting using the CellDrop Trypan Blue Nuclei app. The Trypan Blue dye enters all of the successfully extracted nuclei (circled in red) and stains them, giving them a dark appearance. The intact cells (circled in green) that were not successfully lysed exclude the Trypan Blue dye and appear white.
The numbers of nuclei and intact cells are used to quantify the extraction efficiency for a sample to ensure that enough nuclei are available for the subsequent experiments.
Conclusion
Quality control is an important step in preparing nuclei suspensions for downstream applications. Single-cell sequencing procedures such as those employed by 10X Genomics® rely on isolated nuclei for the technology to appropriately detect expression differences in a cellular population. The CellDrop Automated Cell Counter used in conjunction with AO/PI fluorescence viability dyes provides easy, rapid, and accurate sample analysis. Learn more about counting isolated nuclei on the CellDrop.
Click the button below to view a list of nuclei count settings for various tissue types.
14-AUG-2024
10x Genomics® is a registered trademark of 10x Genomics, Inc. and is used for identification and references purposes only. DeNovix, DeNovix products, and this website are not endorsed or authorized by, or in any way affiliated with 10x Genomics, Inc.
gentleMACS™ Dissociator and gentleMACS™ Octo Coolers are registered trademarks of Miltenyi Biotec, and are used for identification and references purposes only. DeNovix, DeNovix products, and this website are not endorsed or authorized by, or in any way affiliated with Miltenyi Biotec.