Written by Famatta Perry, PhD
How Do You Calculate Protein Concentration for Unknown Samples?
The concentration of unknown protein is calculated by using the protein E1%, a derivative of the molar absorptivity. That is, the protein E1% is used in the Beer’s Law formula to replace the molar absorption coefficient (ε).
Ultraviolet-visible (UV-Vis) spectroscopy is a staple tool for direct protein quantification and is particularly valuable for protein concentration measurements. Spectrophotometry is, in fact, one of the most widely used analytical methods due to its versatility across scientific disciplines for the analysis of molecules and compounds—even in unknown samples.
How is Protein Concentration Measured?
One spectrophotometric method used to accurately calculate the concentration of proteins in solution uses the amount of light absorbed by the protein at 280 nm (A280). However, this method depends on the user knowing the sample’s molar absorption coefficient (also known as molar absorptivity or εmolar),
A molar absorptivity is not known for all proteins, and even when it is, variables such as pH, Ionic strength, or buffer properties can affect the amount of light absorbed.1, 2, 3
In a lab setting, it is also not always the case that the isolated protein(s) are known. Thus, raising the question, how is protein concentration calculated for unknown samples? The answer: E1%.
The objective of this article is to explain the concept of protein E1%. That is, its utilization, presentation and derivation.
What is E1% Protein?
Protein E1% is a derivative of the molar absorptivity that is used in the Beer’s Law formula instead of the molar absorption coefficient (εmolar) for easier calculations and practical conversions. The protein E1% is the absorbance value of 1 gram of a protein in a 100 mL solution, yielding a 1% w/v solution concentration in a standard 1 cm pathlength.1, 2, 3
Thus, the unit of protein E1%, also known as percent solution extinction coefficient (εpercent), is g/100mL or 10 mg/mL.1, 2, 3
Using Beer’s Law, Aλ = ε C L, where Aλ is absorbance, L is pathlength at a 1 cm equivalent, and ε is the molar absorption or extinction coefficient, a protein’s concentration (C) can be easily quantified by measuring its absorbance at a known pathlength.
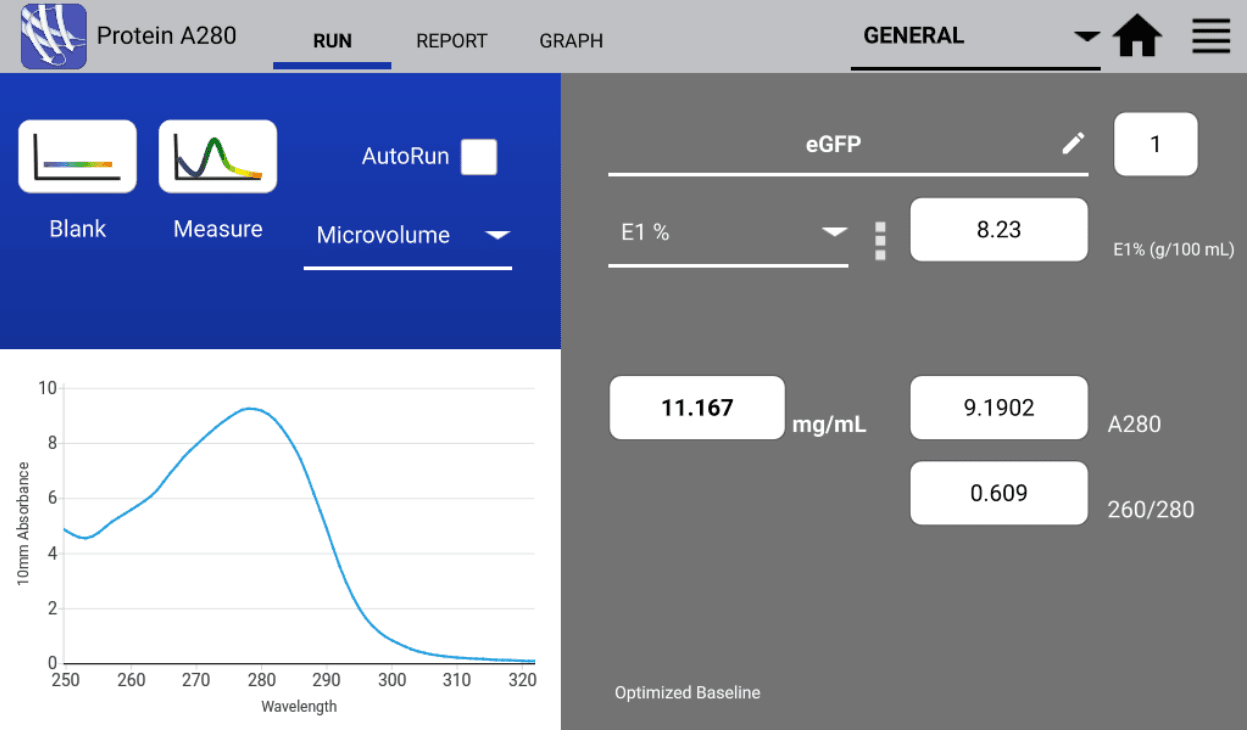
For example, the protein E1% of eGFP at 280 nm is 8.23. This value can then be used to calculate the mg/mL concentration of eGFP from a measured absorbance value of 1.
Note: An adjustment factor of 10 in the equation is used as a conversion factor from percent concentration (g/100 mL) to concentration (mg/mL).
DeNovix DS-Series Spectrophotometers provide a variety of products and user-friendly apps that allows for the customizable calculation of proteins’ and peptides’ concentrations. The Protein A280 app allows users to enter the known protein E1% value and provides the concentration of the sample in mg/mL upon measurement.
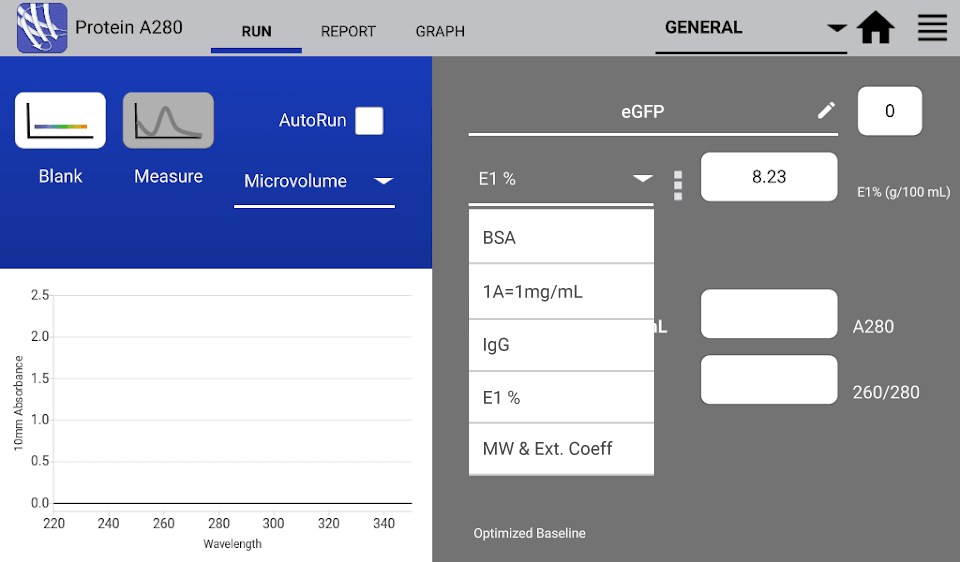
If εpercent value is unknown or not provided, the εmolar can be used to derive the εpercent to easily calculate concentration in mg/mL for practical lab applications. The Formula Methods app can be customized to calculate both the εmolar and the εpercent and provide the concentration of the sample in real time. See the DS-11 Series User Guide for details about using the custom formula options in the Formula Methods app.
You can learn more about different methods of protein quantification by browsing our technical notes. Or, if you have specific questions about measuring protein concentrations with a DeNovix Spectrophotometer, contact a member of the team today.
References
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995 Nov;4(11):2411-23. doi: 10.1002/pro.5560041120. PMID: 8563639; PMCID: PMC2143013.
- Practical Handbook of Biochemistry and Molecular Biology, Fasman, D.G., Ed. (1992). CRC Press, Boston.
- Gill, S.C. and von Hippel, P.H. (1989). Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-26.

Famatta Perry, PhD
DeNovix Application Scientist
Famatta joined the DeNovix team in 2023 after completing her PhD in Animal and Food Science, with a focus on nutritional immunology and immunometabolism, from University of Delaware. At DeNovix, she contributes to product development and application support with expertise in biological assay development and machine learning applications. Outside of work, Famatta enjoys trying new foods, experiencing different cultures through food, and anything related to anime.
Speak to a DeNovix Scientist
Have an application question, or want to learn more about our products? Click the button below to schedule a call with our team of expert application scientists!